Sarah’s previous publications relevant to work in the lab:
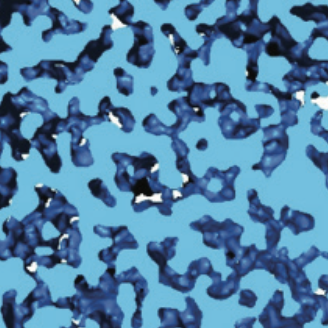
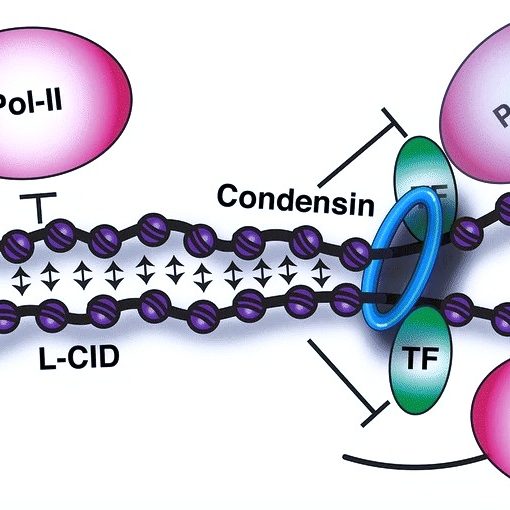
Local chromatin fiber folding represses transcription and loop extrusion in quiescent cells Swygert, S.G., Lin, D., Portillo-Ledesma, S., Lin, P.-Y., Hunt, D.R., Kao, C.-F., Schlick, T., Noble, W.S., and Tsukiyama T. eLife, 2021 Spotlight
Unraveling quiescence-specific repressive chromatin domains Swygert, S.G. and Tsukiyama T. Current Genetics, 2019
Condensin-dependent chromatin compaction represses transcription globally during quiescence Swygert, S.G., Kim, S., Wu, X., Fu, T., Hsieh, T.-H., Rando, O.J., Eisenman, R.N., Shendure, J., McKnight, J.N., and and Tsukiyama T. Molecular Cell, 2019 Spotlight
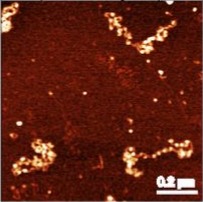
SIR proteins create compact heterochromatin fibers Swygert, S.G., Senapati, S., Bolukbasi, M.F., Wolfe, S.A., Lindsay, S., and Peterson, C.L. Proceedings of the Natural Academy of Sciences, 2018
Preparation and analysis of Saccharomyces cerevisiae quiescent cells Spain, M.M., Swygert, S.G., and Tsukiyama, T. Methods in Molecular Biology: Cellular Quiescence, 2017
Nucleosome-like, single-stranded DNA (ssDNA)-histone octamer complexes and the implication for DNA double strand break repair Adkins, N.L., Swygert, S.G., Kaur, P., Niu, Hengyao, Grigoryev, S.A., Sung, P., Wang, H., Peterson, C.L. Journal of Biological Chemistry, 2017
Solution-state conformation and stoichiometry of yeast Sir3 heterochromatin fibres Swygert, S.G., Manning, B.J., Senapati, S., Kaur, P., Lindsay, S., Demeler, B., Peterson, C.L. Nature Communications, 2014
Chromatin dynamics: Interplay between remodeling enzymes and histone modifications Swygert, S.G., and Peterson, C.L. Biochimica et Biophysica Acta- Gene Regulatory Mechanisms, 2014