How does the conformation of chromatin regulate biological processes?
Although changes in chromatin conformation and mutations in the factors that regulate it have been linked to everything from developmental disorders to aging to cancer, the complexity of chromatin conformations has made determining the specific functions of specific structures in the cell challenging. For this reason, many of the structural mechanisms underlying the regulation of transcription, replication, and DNA repair remain a mystery.
Understanding the relationship between chromatin conformation and its functions requires methods capable of solving chromatin structure at high resolution in cells, while at the same time monitoring processes taking place on the underlying DNA. To address this, we use a genomics method called Micro-C, which generates single-nucleosome resolution maps of all chromatin interactions in the cell. By combining Micro-C with other genomics and microscopy methods, we can piece together the mechanisms by which chromatin conformational changes regulate nuclear processes. As population-based genomics experiments cannot directly determine structure, we also reconstitute relevant chromatin conformations biochemically to link physiological output to structural changes.
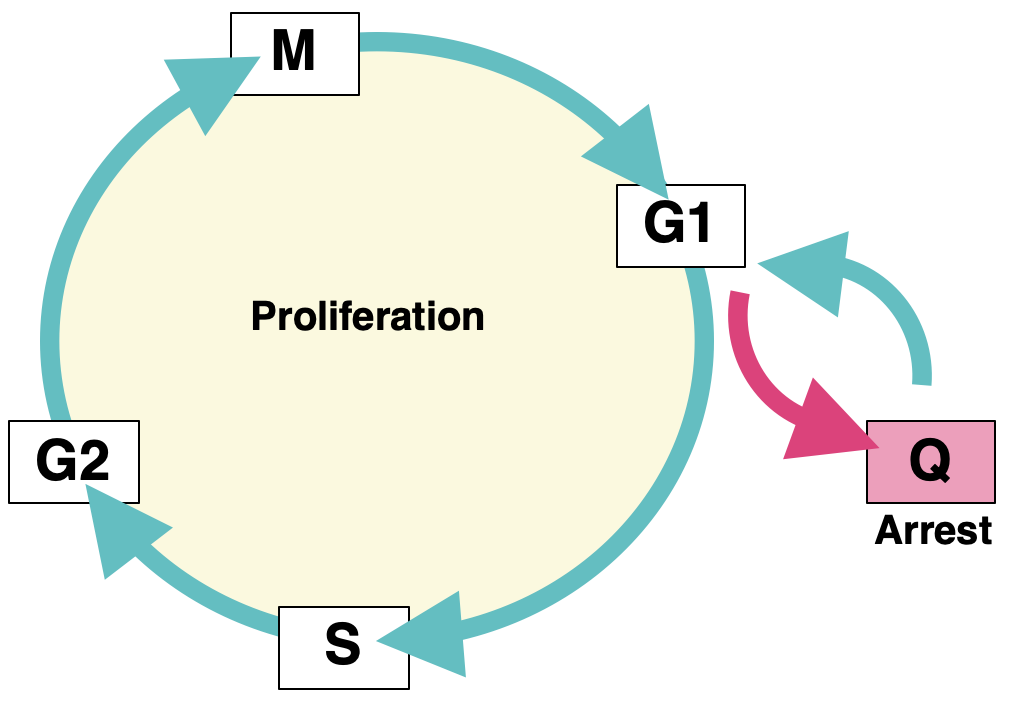
Quiescent yeast are an ideal model for linking chromatin structure to function.
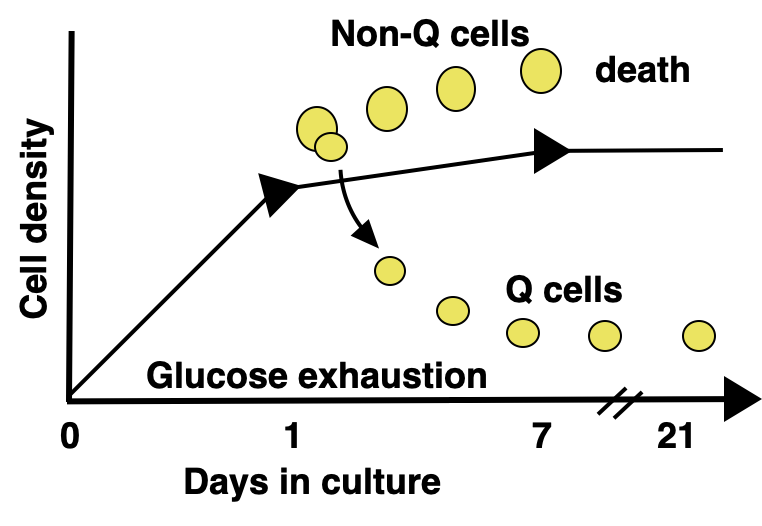
Achieving true single-nucleosome resolution by Micro-C requires extremely deep sequencing to cover every nucleosome in the cell. Additionally, linking chromatin conformation to function requires studying a system that is both biologically and genetically perturbable. Our model system of choice is budding yeast in quiescence. Quiescence is a state in which the cell exits the cell cycle in response to external cues and enters a long-lived and stress-resistant program. Quiescent cells undergo a plethora of profound physiological changes, including widespread transcriptional repression and dramatic chromatin rearrangements. Fascinatingly, quiescent cells are distinct from other G0 states in their ability to exit quiescence and resume cycling. Although yeast cells take several days to enter quiescence (we tend to study them at the one-week mark), cells can exit quiescence in less than five minutes. By studying quiescent yeast, we are able to take advantage of the small S. cerevisiae genome to maximize our sequencing power, study cells undergoing vast chromatin conformational changes directly linked to functional transcriptional changes, and make use of the magic of yeast genetics to perturb any factors of interest. As quiescence is relevant to a variety of biological processes, including lymphocyte activation, wound healing, and cancer recurrence, an added bonus to our research is that we have the ability to uncover both basic regulatory mechanisms and health-relevant quiescence mechanisms.
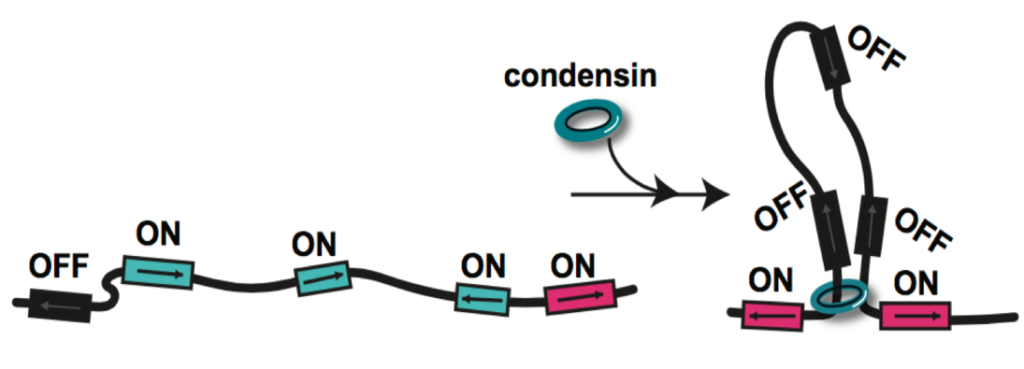
We previously identified chromatin loop domains in quiescent cells formed by the condensin complex. These structures closely resemble mammalian loop domains, often called topologically associated domains (TADs), which are formed by interactions between cohesin and the transcription factor CTCF through a process called loop extrusion. Although TADs have been linked to the regulation of transcription, replication, and repair, the function of the majority of these structures has yet to be uncovered. In contrast, condensin-dependent quiescent domains have a strong functional connection to transcriptional repression during quiescence, as removal of condensin during quiescence entry leads to the transcriptional de-repression of about twenty percent of all genes in the yeast genome.
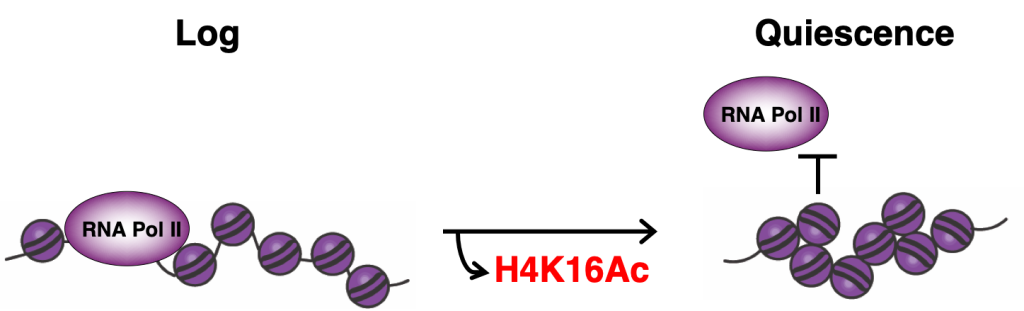
We have also found that quiescent cells compact their chromatin at the local fiber level. This compaction requires a basic patch in the histone H4 tail that becomes deacetylated during quiescence entry by widespread targeting of the histone deacetylase complex Rpd3. Substitutions of residues in the H4 basic patch during quiescence leads to chromatin decompaction and the transcriptional de-repression of about half of all genes. Intriguingly, chromatin fiber decompaction also appears to affect condensin loop formation, perhaps by removing a physical barrier to loop extrusion. These results demonstrate that the conformation of the local chromatin fiber has an important regulatory function in cells.
Current projects in the lab:
How is condensin targeted to chromatin loop domains during quiescence entry, and how does is repress transcription?
During quiescence entry, condensin relocates from its positions at centromeres, the rDNA, and tRNA genes to bind to the promoters of coding genes. Current projects seek to reveal the mechanisms underlying condensin’s migration through the genome. By studying what drives condensin to bind its targets during quiescence, we intend to gain insight into the way in which chromatin loop domains repress transcription.
How does the conformation of the chromatin fiber affect loop extrusion?
One of the biggest mysteries in the genome organization field is how complexes like cohesin and condensin extrude chromatin loops when physiological chromatin is rife with potential barriers. Our previous results suggest that chromatin fiber compaction may impede loop extrusion. By exploring the relationship between chromatin fiber conformation and chromatin loop domains, we will uncover the underlying mechanisms of both types of structures.
How can we link Micro-C data to chromatin structure?
Although Micro-C provides an extremely high-resolution view of chromatin conformation in the cell, we do not yet understand how to interpret Micro-C contact maps to obtain structural information. By completing Micro-C on biochemically reconstituted chromatin fibers of known structure, we hope to generate a key to the Micro-C code that can be used to link contact maps to chromatin structure.
What other architectural factors are involved in regulating chromatin during quiescence?
It is likely that we have only scratched the surface of factors involved in the dramatic rearrangement of chromatin that occurs during quiescence. What other factors are involved in altering chromatin structure during this mysterious state, and what are their functions in the cell?